

REF.1. Model of the structure of the head-to-tail NBD1:NBD2 heterodimer of human CFTR
Reference: Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: detailed sequence analysis and three-dimensional modeling of the heterodimer. Callebaut I, Eudes R, Mornon JP, Lehn P. Cell Mol Life Sci. 2004 Jan;61(2):230-42.
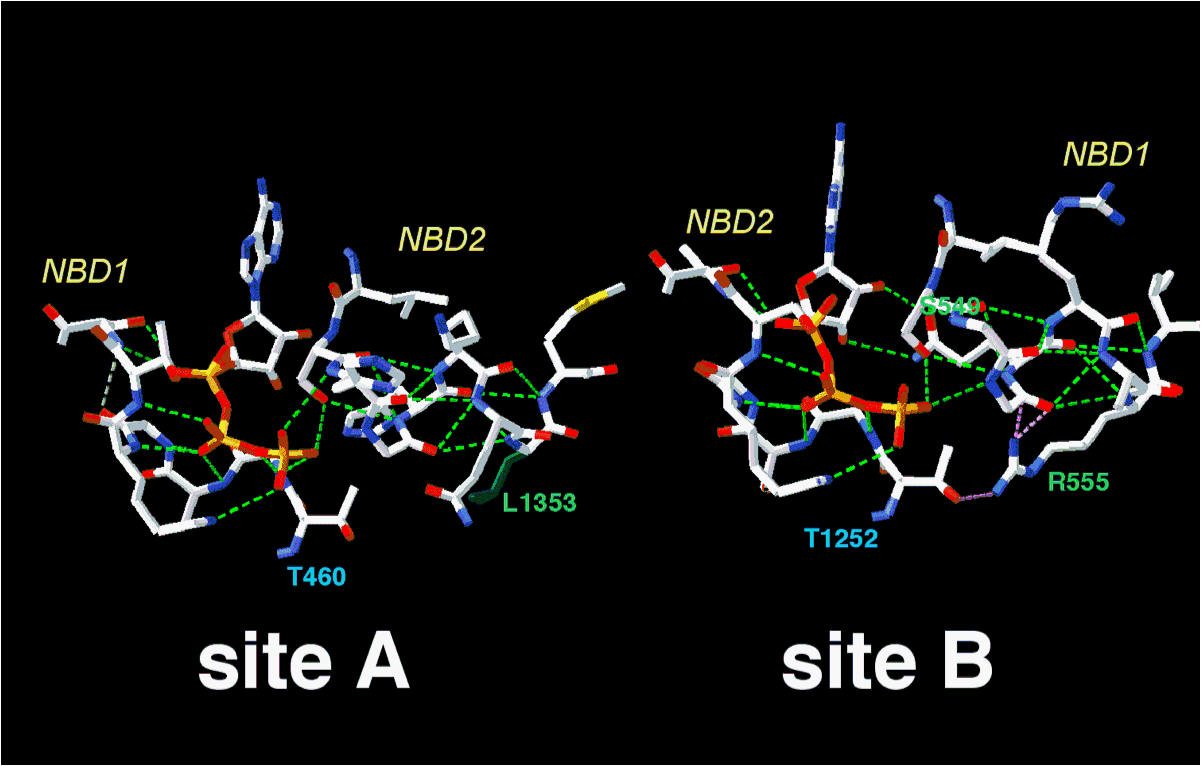
The model of the NBD1 structure within the NBD1:NBD2 heterodimer fits well the experimental structure of mouse NBD1, published at the same time (Lewis et al. 2004, EMBO J 23:282-293). However, relative to the NBD1 isolated 3D structure, our model allowed to predict important features of the NBD1:NBD2 heterodimer interface,which contain the ATP binding sites. For instance, as illustrated below, it can be used to predict and analyze the hydrogen bond network existing in the non conventional (site A) and conventional (site B) ATP binding sites. Among residues which were predicted to interact, the pair R555 (NBD1)/ T1252 (NBD2) was demonstated by Vargani et al. (Nature 2005 433: 876-880) to interact when the channel is open or in the transition state.
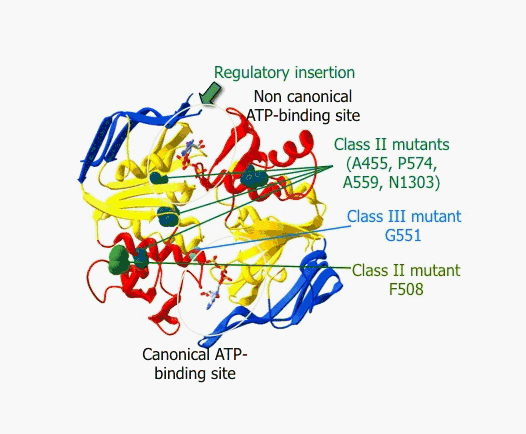
REF.2. Structural classification of mutations affecting the NBD1:NBD2 heterodimer of human CFTR
Reference: Nucleotide binding domains of human CFTR: a structural classification of critical residues and disease-causing mutations. Eudes R, Lehn P, Férec C, Mornon JP, Callebaut I. Cell Mol Life Sci. 2005 Sep;62(18):2112-23.
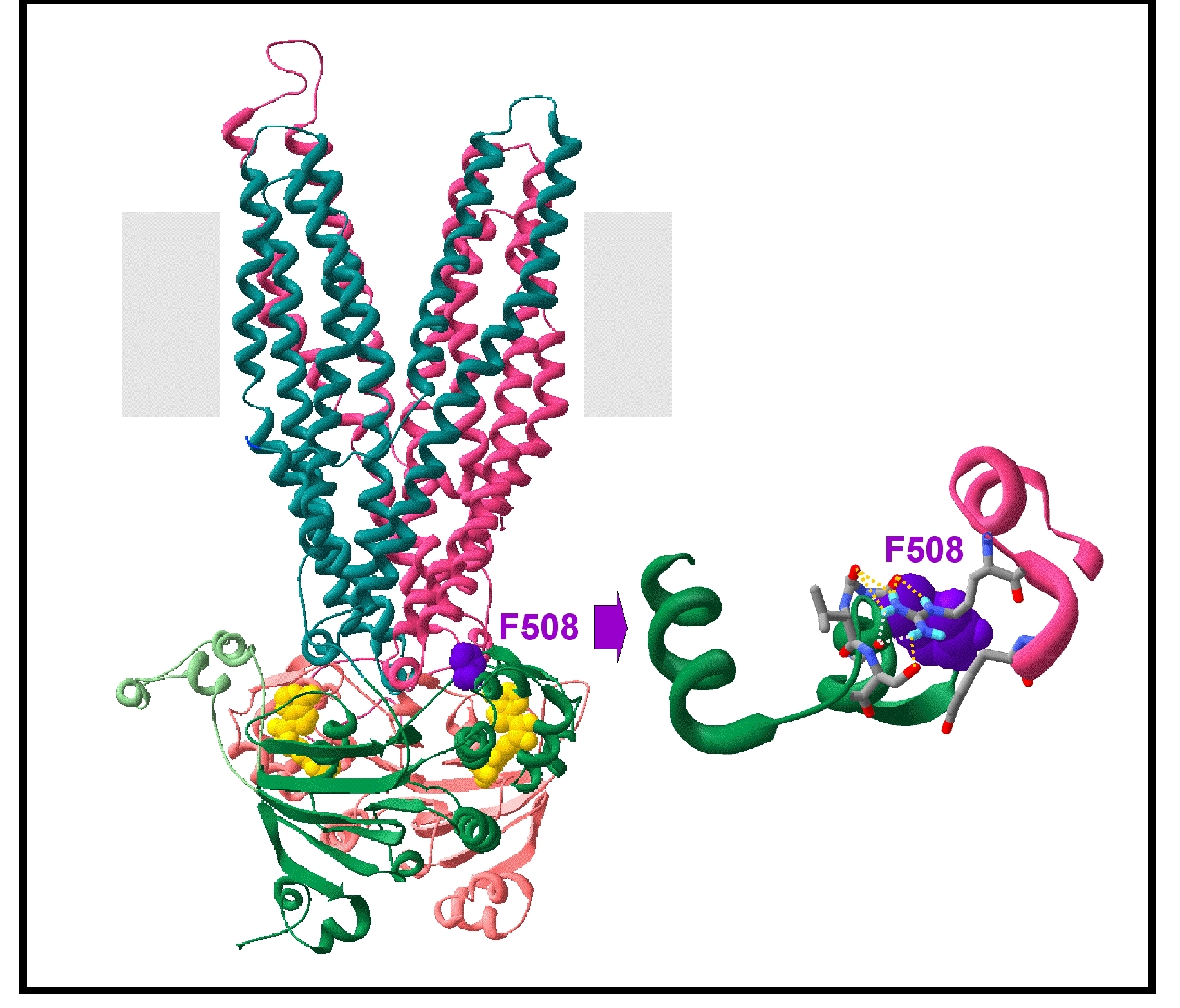
REF.3. Model of an outward-facing conformation of the MSDs:NBDs assembly (based on the Sav1866 3D structure)
Reference: Atomic model of human cystic fibrosis transmembrane conductance regulator: membrane-spanning domains and coupling interfaces. Mornon JP, Lehn P, Callebaut I. Cell Mol Life Sci. 2008 Aug;65(16):2594-612.
Coordinates of the model (pdb file)
This model was made on the basis of the whole structure of Sav1866, thus preserving important features of the domain interfaces. Noticeably, it allowed to highlight important features of the channel pore, as well as of the coupling interfaces between NBDs and MSDs, which include F508 (contacts between NBD1 and the MSD intracellular loop 4 (ICL4), see figure below).
REF.4. Model of an inward-facing conformation of the MSDs:NBDs assembly (based on the MsbA 3D structures)
Reference: Molecular models of the open and closed states of the whole human CFTR protein. Mornon JP, Lehn P, Callebaut I. Cell Mol Life Sci. in press.
Coordinates of the model in the outward-facing conformation (pdb file)
Coordinates of the model in the inward-facing conformation (pdb file)