|
• Background Link to CV with Publications
Since 1996, her research activities have been taking place at the
intersection between informatics, mathematics, physics and biology, with a
focus on conception and implementation of algorithms for biomedical signal
and image processing.
Her first research activities (1996-1999, University of
Belgrade, Serbia) were in biomechanics modeling and signal
processing for control of walking in paraplegic subjects. They evolved
towards multidimensional biomedical data processing during her PhD thesis (1999-2003,
Swiss Federal Institute of Technology in Lausanne - EPFL, Switzerland), through development of image processing
algorithms for computer-assisted surgery and structural biology. During her
postdoc training (2004-2008, University Pierre
and Marie Curie, IMPMC-UMR 7590, Paris, France), she specialized in
electron-microscopy image processing for three-dimensional reconstruction
of biological macromolecular complexes. She obtained a Tenured Associate Scientist position at the French National
Centre for Scientific Research – CNRS in 2008,
a Research Director Habilitation from the Life
Sciences Department of the University Pierre and Marie
Curie - UPMC in 2015, and a CNRS
Research Director position in 2019.
• Focus
Development of algorithms and software that combine image analysis, molecular mechanics simulation, and artificial intelligence to enable full exploration of biomolecular conformational dynamics by in vitro and in situ cryo electron microscopy and cryo electron tomography. These methods are used to study biomedically important molecular complexes in collaboration with experimentalists. For highlights on this research, see below.
• Team members (since 2019)
Current :
Eya Abid, PhD student (Dec 2023 - Present)
Florčne Feyzi, PhD student (Feb 2024 - Present)
Jing Wang, Master intern (Mar 2024 - Present), Sorbonne University, Paris
Shun Robert, Master intern (Mar 2024 - Present), Paris-Saclay University, Orsay
Former :
Mohamad Harastani,
PhD student (Oct 2019 - Oct 2022), currently Postdoc at IGBMC, Illkirch
Ilyes Hamitouche,
PhD student (Oct 2019 - Mar 2023), currently Postdoc at Institut Curie, Paris
Rémi Vuillemot, PhD student (Oct 2020 - Oct 2023), currently Postdoc at Grenoble Alpes University
Alex Mirzaei, Postdoc (Jun 2020 - Nov 2021)
Guchao Zeng, Master intern (May 2023 - Oct 2023), CentraleSupélec, Gif-sur-Yvette
Domique Rado Rakoto, Master intern (May 2023 - Oct 2023), CY Tech, Cergy Pontoise
• Jobs
We are always looking for
enthusiastic and creative Master, PhD, and Postdoc
candidates with an outstanding background in artificial intelligence,
computer vision, image processing, data science, or related fields, wishing
to work on projects on the frontiers of molecular biosciences and software
engineering. Interested candidates should contact Slavica Jonic with a cover letter, CV, and email
addresses of 1-2 references.
• Funding
ANR, CNRS, Sorbonne University, GENCI
• Highlights
We are developing ContinuousFlex [1], a software package containing methods for extracting information on continuous conformational
flexibility of biomolecular
complexes from single particle cryo-EM images and
cryo-ET subtomograms.
The open-source code and the installation instructions can be found at GitHub and PyPI. It is also available as a plugin for Scipion.
Methods available in ContinuousFlex:
- HEMNMA: Hybrid Electron Microscopy Normal Mode Analysis
method to interpret heterogeneity of a set of single particle cryo-EM images in terms of continuous
macromolecular conformational transitions, based on normal mode analysis [2-4]
- StructMap: Structural Mapping method to interpret
heterogeneity of a set of single particle cryo-EM
maps in terms of continuous conformational transitions, based on normal mode analysis [5]
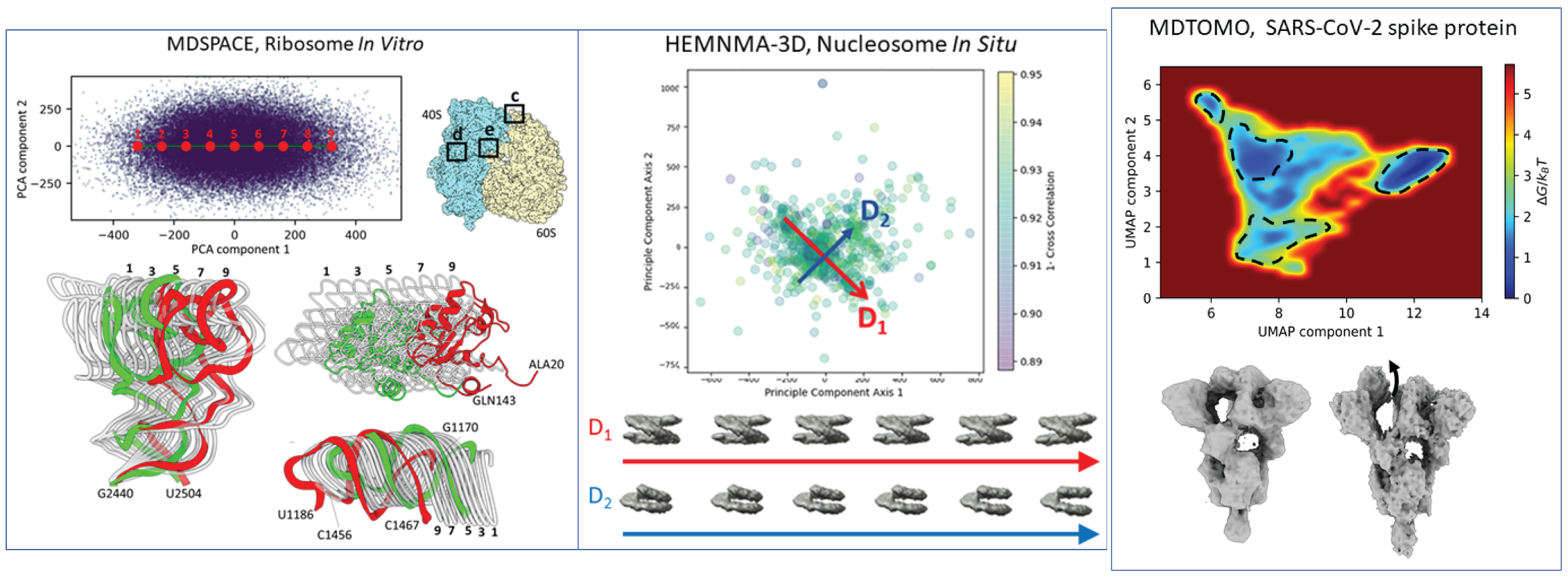
- HEMNMA-3D: Extension of HEMNMA to continuous conformational variability analysis of macromolecules in cryo-ET subtomograms (in vitro and in situ) [6]
- TomoFlow: Method for analyzing continuous conformational variability of macromolecules in cryo-ET subtomograms (in vitro and in situ) based on 3D dense optical flow [7]
- NMMD: Method for flexible fitting of atomic structures into cryo-EM maps using a combination of Normal Mode (NM) analysis and Molecular Dynamics (MD) simulations implemented in GENESIS [8]
- DeepHEMNMA: A deep learning extension of HEMNMA [9]
- MDSPACE: Method for extracting atomic-resolution landscapes of continuous conformational variability of biomolecules from cryo-EM single particle images based on a new 3D-to-2D flexible fitting method, which uses Molecular Dynamics (MD) simulation in combination with normal modes, and is embedded in an iterative conformational-landscape refinement scheme [10,15]
- Data synthetis tools: HEMNMA and HEMNMA-3D protocols additionally provide tools for synthesizing noisy and CTF-affected single particle cryo-EM images (HEMNMA) and noisy, CTF and missing wedge affected cryo-ET subtomograms (HEMNMA-3D) with flexible or rigid biomolecular conformations, for several types of conformational distributions, from a given atomic structure or an EM map. One part of the noise is applied on the ideal projections before and the other after the CTF, as described in [11-12].
- MDTOMO: Method for extracting atomic-resolution landscapes of continuous conformational variability of biomolecules from cryo-ET subtomograms, which uses Molecular Dynamics (MD) simulation in combination with normal modes, and is embedded in an iterative conformational-landscape refinement scheme [13,15]
Applications of ContinuousFlex methods:
- Nucleosome studies by in situ cryo-ET [6,7]
- Yeast 70S ribosomes studies by single-particle cryo-EM [9,10]
- SARS-CoV-2 spike studies by cryo-ET [13]
- ATPase p97 studies by single-particle cryo-EM [16]
- HER2-trastuzumab-pertuzumab studies by single-particle cryo-EM [17]
Online lectures:
- Shapes seminar series, Paris,
Feb 7th, 2024 [YouTube]
-
From cryo-EM data to continuous conformational landscapes
- ARC Centre for Cryo-EM of Membrane Proteins, Melbourne, Australia,
November 4th, 2022 [YouTube]
-
DeepHEMNMA for analyzing continuous conformational heterogeneity in single-particle cryo-EM images
- Instruct Image Processing Center (I2PC) seminar series,
April 15th, 2021 [YouTube]
-
HEMNMA and HEMNMA-3D for
in vitro and in situ studies
of continuous conformational variability of macromolecular
complexes, ContinuousFlex plugin of Scipion
- One World Cryo-EM seminar series, March 24th, 2021 [YouTube]
- Combining normal mode analysis,
image analysis, and deep learning for in vitro and in
situ studies of continuous conformational variability of macromolecular
complexes
Selected publications:
[1] Harastani M, Vuillemot R, Hamitouche I, Moghadam NB, Jonic S: ContinuousFlex: Software package for analyzing continuous conformational variability of macromolecules in cryo electron microscopy and tomography data. J Struct Biol. 2022;214:107906. [Journal][HAL]
[2] Jin Q, Sorzano CO,
de la Rosa-Trevin JM, Bilbao-Castro JR,
Nunez-Ramirez R, Llorca O, Tama F, Jonic S: Iterative elastic 3D-to-2D alignment
method using normal modes for studying structural dynamics of large
macromolecular complexes. Structure 2014, 22:496-506. [Open-access]
[3] Jonic S: Computational
methods for analyzing conformational variability of macromolecular
complexes from cryo-electron microscopy images.
Curr Opin Struct Biol 2017, 43:114-121. [Journal] [Author’s version]
[4] Harastani M, Sorzano CO, Jonic S: Hybrid Electron Microscopy Normal Mode
Analysis with Scipion. Protein Sci 2020,
29:223-36. [Open-access]
[5] Sanchez Sorzano CO, Alvarez-Cabrera
AL, Kazemi M, Carazo JM, Jonic
S: StructMap:
Elastic Distance Analysis of Electron Microscopy Maps for Studying
Conformational Changes. Biophys J 2016, 110:1753-1765.
[Open-access]
[6] Harastani M, Eltsov M, Leforestier A, Jonic S: HEMNMA-3D:
Cryo Electron Tomography Method Based on Normal
Mode Analysis to Study Continuous Conformational Variability of
Macromolecular Complexes. Front Mol Biosci
2021,8:663121. [Open-access]
[7] Harastani M, Eltsov M, Leforestier A, Jonic S: TomoFlow: Analysis of continuous conformational variability of macromolecules in cryogenic subtomograms based on 3D dense optical flow. J Mol Biol 2021, 167381. [Journal] [Author’s version]
[8] Vuillemot R, Miyashita O, Tama F, Rouiller I, Jonic S: NMMD: Efficient Cryo-EM Flexible Fitting Based on Simultaneous Normal Mode and Molecular Dynamics atomic displacements. J Mol Biol 2022, 167483. [Journal] [Author’s version]
[9] Hamitouche I and Jonic S: DeepHEMNMA: ResNet-based hybrid analysis of continuous conformational heterogeneity in cryo-EM single particle images. Front Mol Biosci 2022 9:965645.[Journal] [Author’s version]
[10] Vuillemot R, Mirzaei A, Harastani M, Hamitouche I, Fréchin L, Klaholz BP, Miyashita O, Tama F, Rouiller I, Jonic S: MDSPACE: Extracting continuous conformational landscapes from cryo-EM single particle datasets using 3D-to-2D flexible fitting based on Molecular Dynamics simulation. J Mol Biol 2023,167951. [Journal]
[11] C.O.S. Sorzano, S. Jonic, R. Núñez-Ramírez, N. Boisset, J.M. Carazo: Fast, robust, and accurate determination of transmission electron microscopy contrast transfer function. J Struct Biol. 2007, 160: 249-262. [Journal]
[12] Jonic S, Sorzano CO, Thevenaz P, El-Bez C, De Carlo S, Unser M: Spline-based
image-to-volume registration for three-dimensional electron microscopy.
Ultramicroscopy
2005, 103:303-317. [Author’s version]
[13] Vuillemot R, Rouiller I, Jonic S: MDTOMO method for continuous conformational variability analysis in cryo electron subtomograms based on molecular dynamics simulations. Sci Rep 2023, 13, 10596. [Open-access]
[14] Harastani M and Jonic S: Methods for analyzing continuous conformational variability of biomolecules in cryo electron subtomograms: HEMNMA-3D vs. traditional classification. BioRxiv 2021 [Open-access]
[15] Vuillemot R, Harastani M, Hamitouche I, Jonic S : MDSPACE and MDTOMO Software for Extracting Continuous Conformational Landscapes from Datasets of Single Particle Images and Subtomograms Based on Molecular Dynamics Simulations: Latest Developments in ContinuousFlex Software Package. Int J Mol Sci 2024. 25, 20 [Open-access]
[16] Valimehr S, Vuillemot R, Kazemi M, Jonic S, Rouiller I: Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling Continuous Conformational Variability of Partially Symmetrical Complexes. Int J Mol Sci 2024, 25, 3371. (2 co-first authors) [Open-access]
[17] Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic S, Mathieu M, Bressanelli S: Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex . J Struct Biol 2024, 216(2): 108095 [Journal]
Published datasets (open-access):
[1] Vuillemot R & Jonic S (2022) Data used in J Mol Biol 167951, 2023 https://doi.org/10.5281/zenodo.7415104
[2] Hamitouche I & Jonic S (2022) Data used in Front Mol Biosci 9: 965645, 2022. https://doi.org/10.5281/zenodo.7051222
[3] Harastani M & Jonic S (2021) Data used in J Mol Biol 434(2):167381, 2022. https://doi.org/10.5281/zenodo.5718820
[4] Harastani M, Eltsov M, Leforestier A, Jonic S (2021) Data used in Front Mol Biosci 8:663121, 2021 https://www.ebi.ac.uk/emdb/EMD-12699 and https://doi.org/10.6019/empiar-10679.
|